This tutorial gives an introduction to the backstaff command line tool and how it can be used to convert and process Bifrost simulation data to visualize it. It also gives a primer on how ParaView can be used to visualize the data.
Table of contents
- Installing the command line tool
- Fetching demo snapshots
- Inspecting a snapshot
- Converting a snapshot to NetCDF
- Simple visualization with ParaView
- Resampling snapshots
- Volume rendering with ParaView
Installing the command line tool
Making sure NetCDF is available
In this tutorial, we will create NetCDF files for visualization, and for that, we need to have the NetCDF library available.
To check if it is available on your system, run
nc-config --prefix
This will print the path to the directory where the library is located if it exists. If the command fails, install the library using (on macOS)
brew install netcdf
or (on Linux)
sudo apt install netcdf
Running the installation script
The Backstaff repository contains an interactive shell script called setup_backstaff that lets you select the desired features, verifies any dependencies, and builds and installs the backstaff binary.
We can download and run it in one go using the following command:
bash <(curl -s https://raw.githubusercontent.com/lars-frogner/Backstaff/main/setup_backstaff)
If you do not have the Rust toolchain installed on your system, you will be prompted to install it. Say yes and follow the instructions.
We can then move on to selecting features.
The feature system allows us to include only the functionality we need when building the binary. By not having unneeded features, we can avoid unnecessary dependencies and reduce the compilation time and executable binary size. You can find descriptions of the available features at https://github.com/lars-frogner/Backstaff/blob/main/README.md#features.
For this tutorial, we will need the statistics and netcdf features, so say yes to include each of these when prompted. If you think you may need additional features in the future, you can include those now as well. You can always re-run the installation script at a later point to change the feature set.
When prompted about link time optimization, you can say no, and you should say no to using a different version than the default. Finally, you may specify the path where the binary should be installed or just accept the default path.
When the installation has finished, you will be asked if you want to set up tab-completion for the backstaff command. If you want that, say yes and follow the instructions. But be aware that this may affect the responsiveness of constructing commands if you include a lot of features.
If you installed backstaff to a directory in your $PATH, you should now be able to execute the binary:
backstaff -h
You should see a help text like this:
backstaff 0.3.0
Lars Frogner <lars.frogner@astro.uio.no>
A flexible toolkit for working with Bifrost simulations
For documentation, see https://github.com/lars-frogner/Backstaff
USAGE:
backstaff [OPTIONS] <SUBCOMMAND>
OPTIONS:
-t, --timing
Display elapsed time when done
-P, --protected-file-types=<EXTENSIONS>...
List of extensions for file types that never should be overwritten automatically
(comma-separated) [default: idl,snap,aux]
-N, --no-protected-files
Allow any file type to be overwritten automatically
-h, --help
Print help information
-V, --version
Print version information
SUBCOMMANDS:
snapshot Specify input snapshot to perform further actions on
create_mesh Create a Bifrost mesh file
Fetching demo snapshots
We will work with a set of small 3D snapshots from a Bifrost simulation in this tutorial. Use the following commands to download the snapshots from the GitHub repository and extract them:
curl -LO https://github.com/lars-frogner/Backstaff/releases/latest/download/demo_snapshots.zip
unzip demo_snapshots.zip
In the extracted demo_snapshots folder, you will find three sets of files representing a snapshot of the simulation at three points in time (snapshots 413, 414 and 415). Each snapshot has a *.idl parameter file containing all metadata for the snapshot in plaintext, as well as a binary *.snap and *.aux file containing the actual values for respectively the primary and auxiliary quantities. There is also a shared *.mesh text file that defines the simulation grid.
Inspecting a snapshot
The first thing we will do is to find some statistics for the density and temperature fields in a snapshot:
backstaff snapshot demo_snapshots/en2431em_413.idl -v inspect -I=r,tg statistics
A backstaff command consists of a chain of subcommands, each with a set of arguments and/or options specified directly after the name of the subcommand. In the above command, the first subcommand is snapshot, which we have given a (required) argument specifying the path to the parameter file of the snapshot to read, followed by the option -v (verbose) to turn on status messages related to reading snapshots.
There are several different subcommands we could have specified after the snapshot subcommand depending on what we wanted to do with the snapshot. In this case, we used the inspect subcommand with the option -I=r,tg, which says to include only the quantities r (density) and tg (temperature) in the inspection. We could also have used the long form --included-quantities=r,tg.
We finished off with the statistics subcommand without any options to let us inspect the default statistics for the quantities.
Here is the output:
Reading parameters from en2431em_413.idl
Reading grid from en2431em.mesh
Detected horizontally regular grid
Reading r from en2431em_413.snap
================================================================================
Statistics for r from snapshot 413 of en2431em
--------------------------------------------------------------------------------
For all r, all x, all y, all z
================================================================================
Number of values: 1000000
Minimum value: 1.8765084e-8 at ( 17.864, 9.944, -9.409) [ 74, 41, 9]
Maximum value: 263.97241211 at ( 2.744, 17.144, 2.445) [ 11, 71, 99]
Average value: 12.518044472
Reading tg from en2431em_413.aux
================================================================================
Statistics for tg from snapshot 413 of en2431em
--------------------------------------------------------------------------------
For all tg, all x, all y, all z
================================================================================
Number of values: 1000000
Minimum value: 1998.7069092 at ( 7.304, 10.184, -1.540) [ 30, 42, 62]
Maximum value: 7.12432150e6 at ( 13.304, 3.224, -14.057) [ 55, 13, 0]
Average value: 7.52753062e5
--------------------------------------------------------------------------------
Converting a snapshot to NetCDF
Since the native format used for Bifrost snapshots is not standard, 3D visualization tools like ParaView and VAPOR don’t know how to interpret the simulation data directly. The solution is to convert the data into a format that they do understand, and one such format is NetCDF (*.nc). Doing this with Backstaff (installed with the netcdf feature) is simple:
cd demo_snapshots
backstaff snapshot en2431em_413.idl write en2431em_413.nc
Here, we used another of snapshot’s subcommands, write. It needs an argument specifying the path of the output file that it should write. It looks at the output file extension to determine which format to write in, which in this case is NetCDF due to the .nc extension.
Both snapshot and write can handle snapshot data in both NetCDF and native (*.idl) format, so we can read in and write out any combination of the two formats.
Simple visualization with ParaView
Loading the data
Let us load en2431em_413.nc into ParaView to have a look at it. In ParaView, click File -> Open, find and double-click on en2431em_413.nc. When asked which reader to use, double-click on the one labeled “NetCDF Reader”.
Before the data is loaded, we should make a couple of tweaks in the Properties panel (usually to the left of the render view). First, uncheck the Spherical Coordinates checkbox, as our data grid uses Cartesian coordinates.
Second, look at the Dimensions dropdown list just above the checkbox. Each list element represents a grid in the NetCDF file. The file contains multiple grids because Bifrost uses a staggered grid, meaning that some quantities are defined in the centers of the grid cells while others are defined on the faces. ParaView assumes that all quantities loaded with a grid are defined at the same location, so the file needs a separate grid for each place in a grid cell quantities can be defined. For example, temperature (like all other auxiliary quantities) is a cell-centered quantity, so to get access to it, we must load the data with the (zm, ym, xm) grid (zm is the array of cell-centered z-coordinates, etc.). If we hit Apply after selecting this grid, all the cell-centered quantities in the file are loaded. If we instead selected (zmdn, ym, xm), all quantities defined in the middle of the bottom face of the grid cells, like pz and bz, would be loaded instead. But let us stick with the cell-centered quantities for now.
In the render view, you should see the outline of the simulation box. We can add axes by checking the Axes Grid checkbox a bit down in the Properties panel. To look at the actual data, we can apply a Filter, which is ParaView’s mechanism for transforming the data into various forms it can visualize.
2D slice
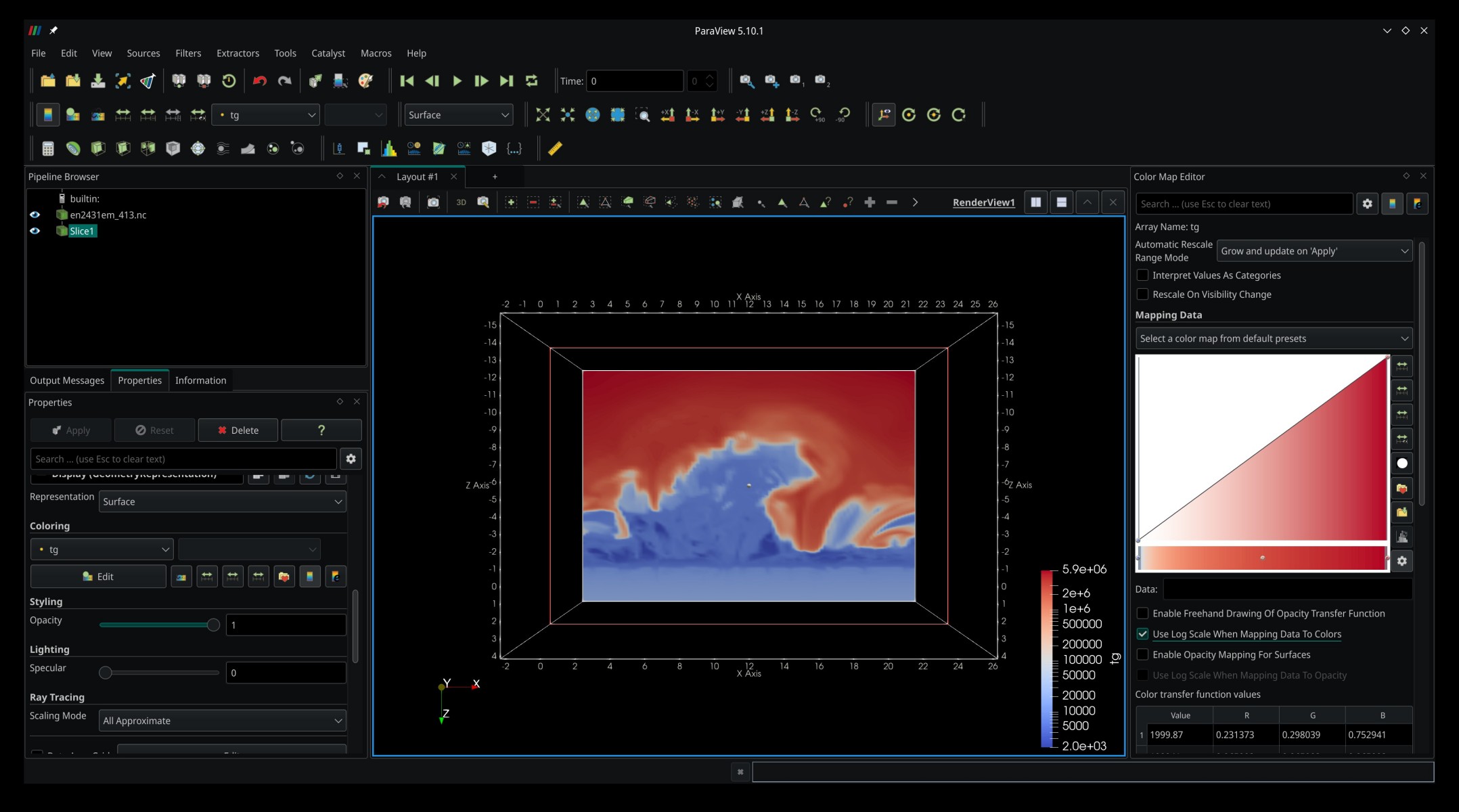
We start by rendering a 2D slice through the simulation box. Click Filters -> Common -> Slice to create a Slice filter. We can adjust the position and orientation of the slice plane either by pulling on the plane or its normal vector in the render view or by using the relevant widgets in the Properties panel. Once happy, hit Apply, and ParaView will render the slice. Disappointingly, it is just plain white. We can fix that by going to the Coloring section of the Properties panel and changing from Solid Color to tg in the dropdown list, which will color the slice according to the local temperature. You can change or adjust the color map in the Color Map Editor (ensure the Slice filter is selected in the Pipeline Browser). You may end up with something that looks like this:
Isosurface
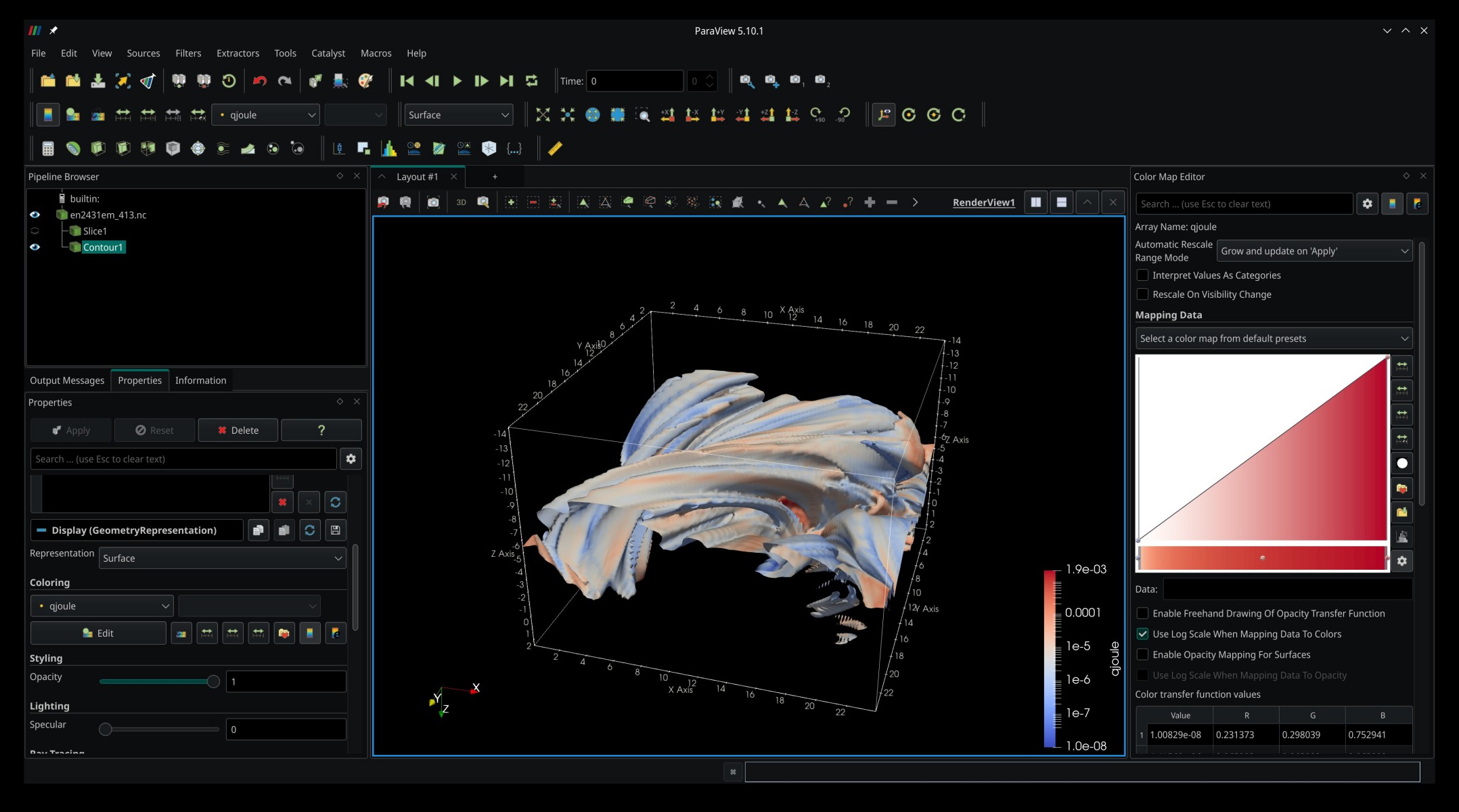
Let us try something a bit more exciting and render an isosurface. Hide our slice plane by clicking on the eye next to Slice1 in the Pipeline Browser. Then, select en2431em_413.nc, as we will apply the new filter to the original data source, not the data outputted from the Slice filter. Click Filters -> Common -> Contour. In the properties of the newly created Contour filter, select tg in the Contour By list, and then add 1e6 in the Isosurfaces box below. Hit Apply and behold the isosurface for 1 MK. You can add as many isosurfaces as you like in the Isosurfaces box. By default, the coloring of the surfaces is based on the quantity we create the contour for, but that can easily be changed by selecting another quantity under Coloring in the Properties panel. Here is what you might get if you color the contour by resistive heating (qjoule):
Resampling snapshots
Resampling to a reshaped grid
The coarse spatial resolution of the demo snapshots (100 grid cells in each dimension) makes our isosurface look a bit jagged. We could make it look smoother by upsampling the snapshots, which gives us a perfect excuse to try out another backstaff subcommand: resample. But instead of conjuring the correct command out of thin air like before, we will now build the appropriate command incrementally.
We know we want to operate on a snapshot, so we start with the snapshot subcommand and add the -h flag to print the help text for snapshot so we can see what further options we have:
backstaff snapshot -h
Here is the output:
backstaff-snapshot
Specify input snapshot to perform further actions on
USAGE:
backstaff snapshot [OPTIONS] <INPUT_FILE> <SUBCOMMAND>
ARGS:
<INPUT_FILE> Path to the file representing the snapshot.
Assumes the following format based on the file extension:
*.idl: Parameter file with associated .snap [and .aux] file
*.nc: NetCDF file using the CF convention (requires the netcdf feature)
OPTIONS:
-r, --snap-range=<FIRST>,<LAST> Inclusive range of snapshot numbers associated with the input
snapshot to process [default: only process INPUT_FILE]
-e, --endianness=<ENDIANNESS> Endianness to assume for snapshots in native binary format
[default: little] [possible values: little, big, native]
-v, --verbose Print status messages related to reading
-h, --help Print help information
SUBCOMMANDS:
inspect Inspect properties of the snapshot
slice Extract a 2D slice of a quantity field in the snapshot
extract Extract a subdomain of the snapshot
resample Create a resampled version of the snapshot
write Write snapshot data to file
We see that resample is available as a subcommand of snapshot, so we should add that to our command. But before adding resample, we must remember to add any arguments and options we want to pass to the snapshot subcommand. In this case we’ll just specify the input file path. This is our new command:
backstaff snapshot en2431em_413.idl resample -h
Notice that we have moved the -h flag to after resample, which will give us the help text for the resample subcommand. In general, the command will do nothing but show a help text if -h is present anywhere in the command. If the -h flag is present for multiple subcommands, only the help text of the first of them will be shown.
Here is the output:
backstaff-snapshot-resample
Create a resampled version of the snapshot
USAGE:
backstaff snapshot <INPUT_FILE> resample [OPTIONS] <SUBCOMMAND>
OPTIONS:
-l, --sample-location=<LOCATION> Location within the grid cell where resampled values should
be specified
[default: original] [possible values: original, center]
--ignore-warnings Automatically continue on warnings
-v, --verbose Print status messages related to resampling
-p, --progress Show progress bar for resampling (also implies `verbose`)
-h, --help Print help information
SUBCOMMANDS:
regular_grid Resample to a regular grid
rotated_regular_grid Resample to a regular grid rotated around the z-axis
reshaped_grid Resample to a reshaped version of the original grid
mesh_file Resample to a grid specified by a mesh file
We see that resample must be followed by a subcommand that specifies what kind of grid to resample to. For our current purpose, we simply want a higher-resolution version of our original grid. The best choice for this is reshaped_grid, which preserves the bounds, orientation and non-uniformity of the original grid. Let us also use the opportunity to make all the quantities cell-centered by including the option --sample-location=center. Finally, we’ll include the -p flag to get progress bars when resampling. The next version of the command then becomes:
backstaff snapshot en2431em_413.idl \
resample -p --sample-location=center reshaped_grid -h
backstaff-snapshot-resample-reshaped_grid
Resample to a reshaped version of the original grid
USAGE:
backstaff snapshot resample reshaped_grid [OPTIONS] <SUBCOMMAND>
OPTIONS:
-s, --shape=<NX>,<NY>,<NZ> Shape of the resampled grid (`same` is original size) [default:
same,same,same]
-c, --scales=<SX>,<SY>,<SZ> Factors for scaling the dimensions specified in the `shape`
argument [default: 1,1,1]
-h, --help Print help information
SUBCOMMANDS:
sample_averaging Use the weighted sample averaging method
cell_averaging Use the weighted cell averaging method
direct_sampling Use the direct sampling method
write Write snapshot data to file
inspect Inspect properties of the snapshot
You can use a subcommand to configure the resampling method. If left unspecified,
sample averaging with the default prameters is used.
We can see the light in the end of the tunnel now. To upsample the snapshot to double resolution in each dimension, we include the option --scales=2,2,2. We could add a subcommand to request a non-default resampling method (cell_averaging or direct_sampling), but it is usually best to stick with the default method, sample_averaging, which works great for any kind of resampling with the expense of being a bit slower. We can then go straight to the write subcommand, giving:
backstaff snapshot en2431em_413.idl \
resample -p --sample-location=center reshaped_grid --scales=2,2,2 \
write -h
Our last help text looks like this:
backstaff-snapshot-resample-reshaped_grid-write
Write snapshot data to file
USAGE:
backstaff snapshot resample reshaped_grid write [OPTIONS] <OUTPUT_FILE>
ARGS:
<OUTPUT_FILE> Path of the output file to produce.
Writes in the following format based on the file extension:
*.idl: Creates a parameter file with an associated .snap [and .aux]
file
*.nc: Creates a NetCDF file using the CF convention (requires the
netcdf feature)
If processing multiple snapshots, the output snapshot number will be
incremented (or appended if necessary) with basis in this snapshot file
name.
OPTIONS:
--overwrite
Automatically overwrite any existing files (unless listed as protected)
--no-overwrite
Do not overwrite any existing files
-I, --included-quantities=<NAMES>...
List of all the original quantities to include in the output snapshot
(comma-separated) [default: all]
-E, --excluded-quantities=<NAMES>...
List of original quantities to leave out of the output snapshot
(comma-separated)
--ignore-warnings
Automatically continue on warnings
-v, --verbose
Print status messages related to writing
-s, --strip
Strip away metadata not required for visualization
-h, --help
Print help information
Like before, we must provide an output file path to write. We still want a NetCDF file, so let’s call it en2431em_upsampled_413.nc. Here is the final command:
backstaff snapshot en2431em_413.idl \
resample -p --sample-location=center reshaped_grid --scales=2,2,2 \
write en2431em_upsampled_413.nc
Running it yields the following output (with a progress bar being completed for each line along the way);
Resampling r
Resampling px
Resampling py
Resampling pz
Resampling e
Resampling bx
Resampling by
Resampling bz
Resampling p
Resampling tg
Resampling nel
Resampling qjoule
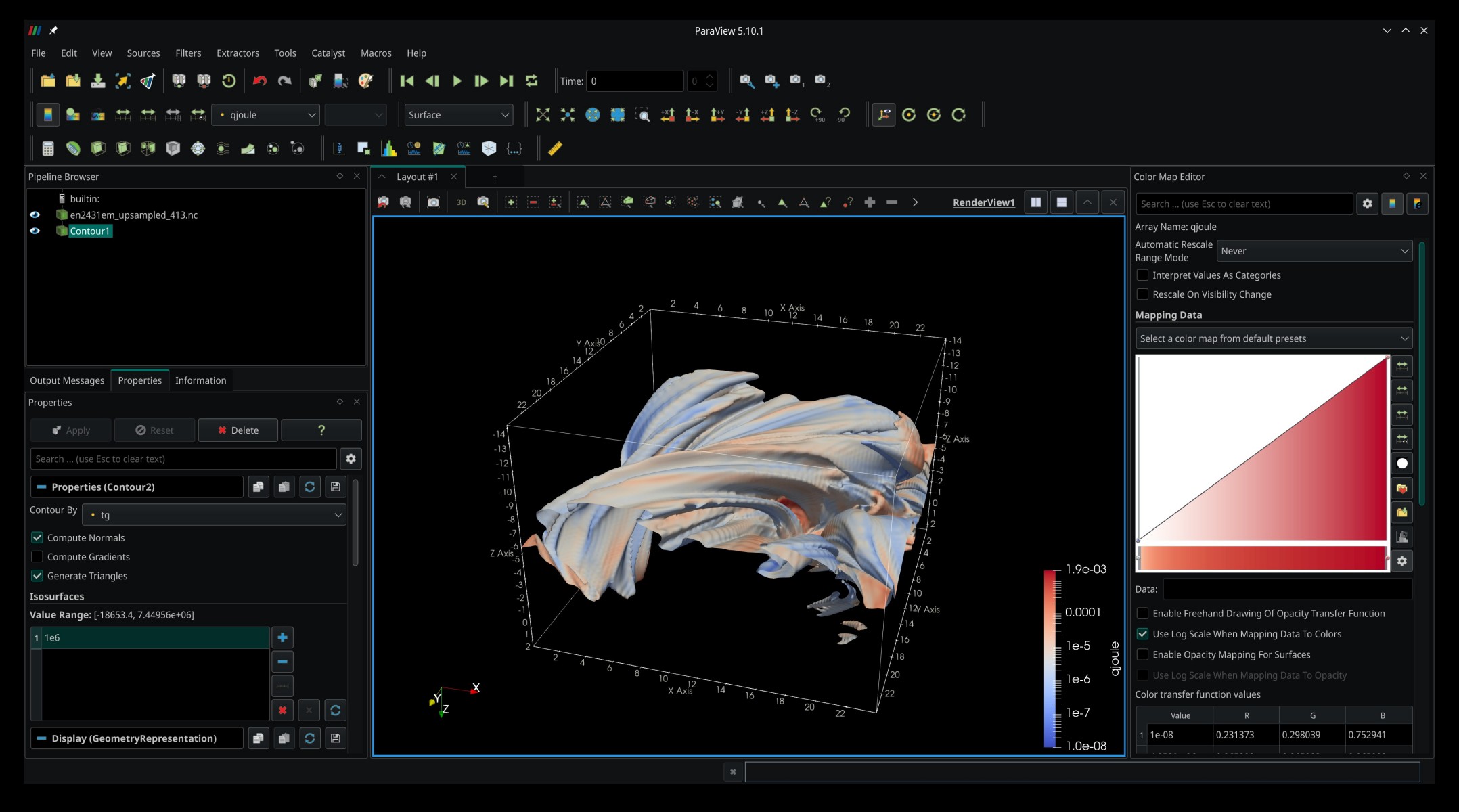
as well as the output file en2431em_upsampled_413.nc. If we load it in ParaView and render an isosurface like before, we get a slightly smoother result:
The upsampling seems to have worked!
NOTE: This example is admittedly somewhat contrived, since we ordinarily wouldn’t want to upsample simulation data for visualization. A more realistic use case for upsampling would be to produce a higher-resolution version of a snapshot for further simulation. The command for that would be identical except for the --sample-location option and that we would use idl as the extension for the output file instead of nc.
Resampling to a regular grid
As we saw in the previous example, the resample subcommand has several subcommands representing different ways in which the snapshot can be resampled. One that is particularly useful for visualization is regular_grid. As the name implies, this subcommand resamples the snapshot to a grid where the grid cell extent is constant along all dimensions. This is different from the grids used in most Bifrost simulations, which tend to have a varying resolution in the z-direction.
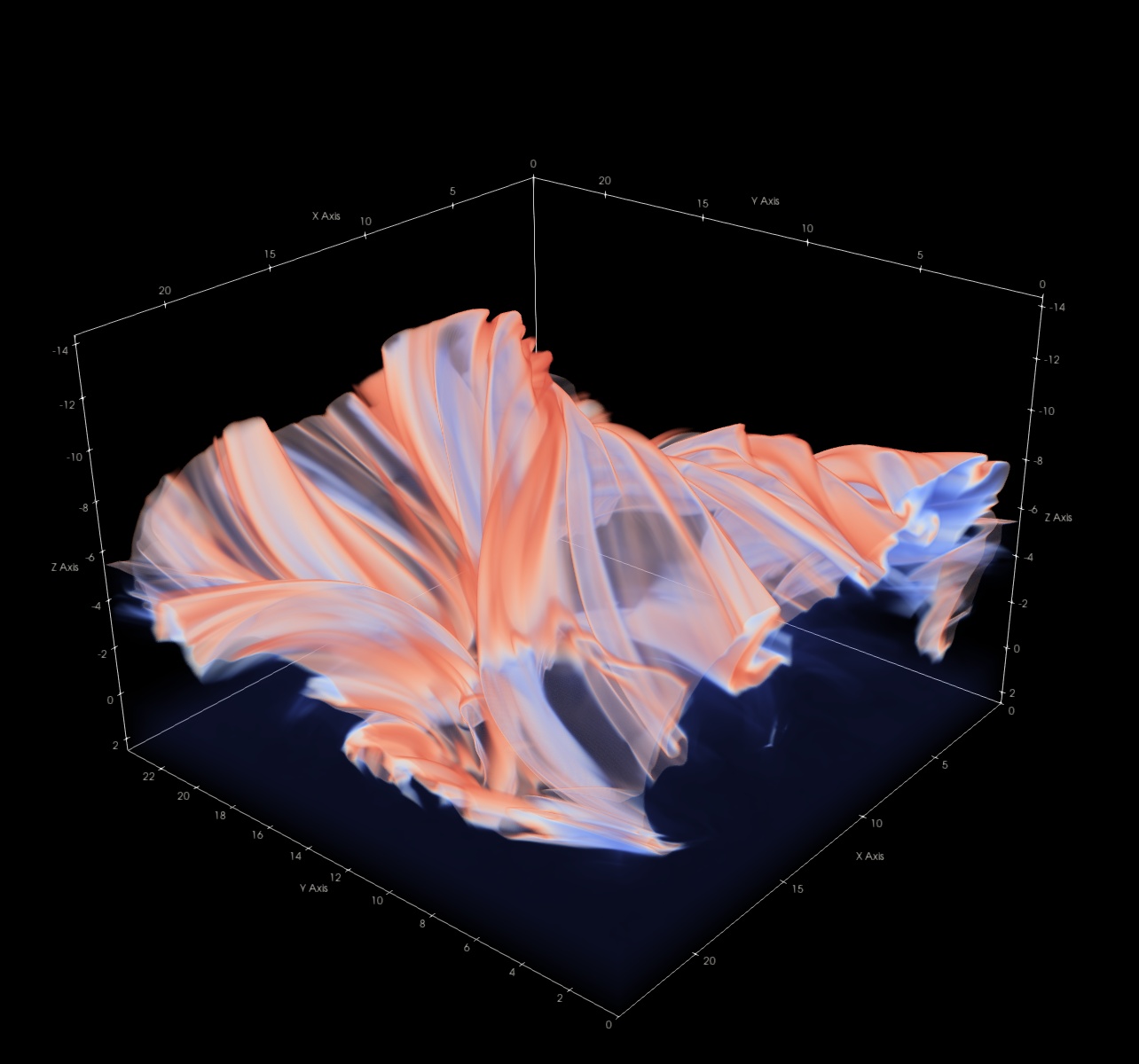
While varying vertical resolution is good for simulations, it causes problems when we want to do interactive volume rendering. Volume rendering is a technique where we assign a specific color and opacity to each grid cell based on the local value of the quantity we want to visualize. By blending the colors together along each line of sight from a given view point, we obtain an image that resembles what we would see if we were looking into a cloud of emissive gas from the view point. Here is an example of a ParaView volume rendering of the transition region temperatures in a snapshot:
Volume rendering can be done very efficiently on a GPU, enabling relatively large fields to be visualized at interactive speeds. Unfortunately, this speed is only attainable if the grid is regular. If not, rendering will be orders of magnitude slower. So if we want to volume render our snapshot, we must resample it first.
To do this with backstaff, we can recycle most of the command we used for upsampling in the previous section. Let us change the output file name to en2431em_regular_413.nc and swap out the reshaped_grid subcommand and its option for the regular_grid subcommand with a -h flag.
backstaff snapshot en2431em_413.idl \
resample -p --sample-location=center regular_grid -h \
write en2431em_regular_413.nc
Recall that as long as there is a a help flag somewhere in the command, the only thing the command will do is to print out the corresponding help text.
backstaff-snapshot-resample-regular_grid
Resample to a regular grid
USAGE:
backstaff snapshot resample regular_grid [OPTIONS] <SUBCOMMAND>
OPTIONS:
-s, --shape=<NX>,<NY>,<NZ> Shape of the resampled grid (`auto` approximates original cell
extent) [default: auto,auto,auto]
-c, --scales=<SX>,<SY>,<SZ> Factors for scaling the dimensions specified in the `shape`
argument [default: 1,1,1]
-x, --x-bounds=<LOWER>,<UPPER> Limits for the x-coordinates of the resampled grid [default:
min,max]
-y, --y-bounds=<LOWER>,<UPPER> Limits for the y-coordinates of the resampled grid [default:
min,max]
-z, --z-bounds=<LOWER>,<UPPER> Limits for the z-coordinates of the resampled grid [default:
min,max]
-h, --help Print help information
SUBCOMMANDS:
sample_averaging Use the weighted sample averaging method
cell_averaging Use the weighted cell averaging method
direct_sampling Use the direct sampling method
write Write snapshot data to file
inspect Inspect properties of the snapshot
You can use a subcommand to configure the resampling method. If left unspecified,
sample averaging with the default prameters is used.
All options here are, well, optional. If we don’t specify otherwise, regular_grid will resample our snapshot to a regular grid with the same bounds and dimensions as the original grid. For our small demo snapshots, this is fine. If our snapshots had a high-resolution grid, say 768 grid cells in each dimension, we would probably want a somewhat downsampled regular grid in order to keep the visualisation interactive. To achieve this we could for instance specify --scale=0.5,0.5,0.5 to get half the original resolution, or --shape=512,512,512 to get exactly 512 grid cells in each dimension. Or we might instead only be interested in looking at some limited subdomain of the simulation box. Then we could specify the lower and upper bounds of the subdomain in each dimension with the -x, -y and -z options, and obtain a snapshot with a regular grid covering only this subdomain.
For this example we will leave regular_grid without options (so remove the -h flag). Instead, let us add some options to the snapshot and write subcommands:
backstaff snapshot en2431em_413.idl --snap-range=413,415 \
resample -p --sample-location=center regular_grid \
write -v -I=r,tg,nel,qjoule en2431em_regular_413.nc
The option we have added to snapshot is --snap-range=413,415. The --snap-range option is very useful for processing a sequence of snapshots from the same simulation in one go. It will make the program read all of the snapshots in the specified range (in this case snaps 413, 414 and 415) one by one and run the other subcommands on each of them. The snapshot numbers in the output files will be incremented accordingly.
For the write subcommand, we have added a -I (long form --included-quantities) option to limit the quantities in the output files to those we might be most interested in volume rendering. We have also added the -v flag to see the command is building up the output files.
The command spits out the following to the terminal:
Writing grid to en2431em_regular_413.nc
Resampling r
Writing r to en2431em_regular_413.nc
Resampling tg
Writing tg to en2431em_regular_413.nc
Resampling nel
Writing nel to en2431em_regular_413.nc
Resampling qjoule
Writing qjoule to en2431em_regular_413.nc
Writing grid to en2431em_regular_414.nc
Resampling r
Writing r to en2431em_regular_414.nc
Resampling tg
Writing tg to en2431em_regular_414.nc
Resampling nel
Writing nel to en2431em_regular_414.nc
Resampling qjoule
Writing qjoule to en2431em_regular_414.nc
Writing grid to en2431em_regular_415.nc
Resampling r
Writing r to en2431em_regular_415.nc
Resampling tg
Writing tg to en2431em_regular_415.nc
Resampling nel
Writing nel to en2431em_regular_415.nc
Resampling qjoule
Writing qjoule to en2431em_regular_415.nc
We are left with three resampled snapshot files: en2431em_regular_413.nc, en2431em_regular_414.nc and en2431em_regular_415.nc.
Volume rendering with ParaView
We will now use Paraview to volume render the snapshots that we resampled to a regular grid in the previous section.
Start by loading the en2431em_regular_*.nc files in ParaView as described in the section on loading data. You will notice that ParaView file browser recognises the three snapshots as belonging to the same group. Opening the whole group instead of individual files will cause the snapshots to be loaded as a time sequence, enabling us to play through the snapshots or select specific times in the time control toolbar near the top of the application.
To produce a volume rendering, find the Representation dropdown list under Display in the Properties panel and change the representation from Outline to Volume. We can see which quantity is being rendered in the dropdown list under Coloring just below. Let us change it to tg to view the temperature.
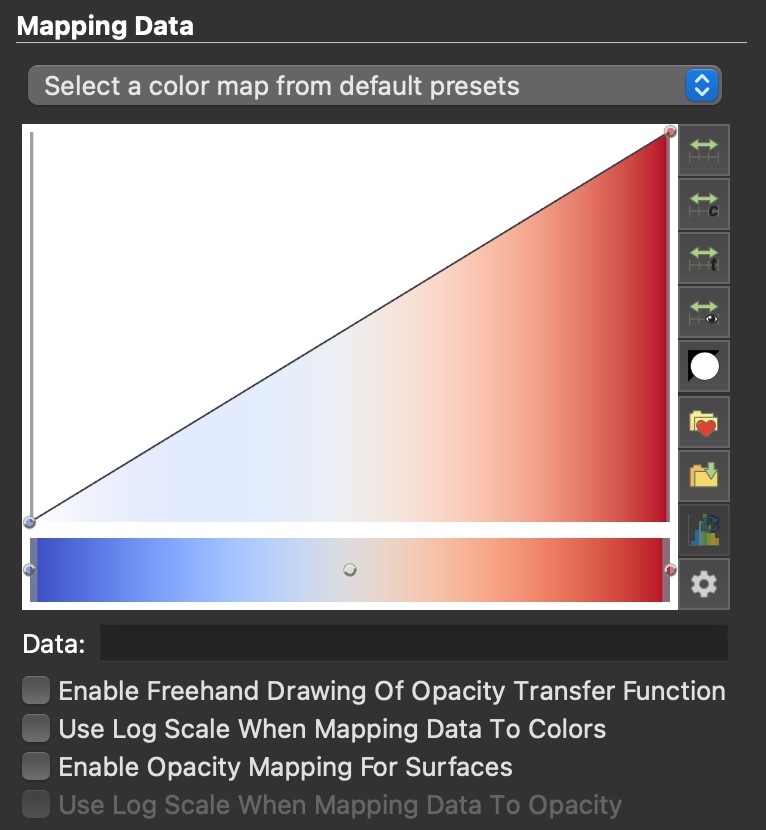
It is usually neccesary to tweak the opacity transfer function in order to get the visualisation we want. The opacity transfer function is responsible for converting the field values to opacities. By manipulating it we can make the values we are uninterested in more transparent to better reveal the values that we do want to see.
The Color Map Editor contains an interactive plot of the opacity transfer function. Its initial form is a linear function that is 0 for the minimum value of the field and 1 for the maximum value. Directly below the graph is a rectangular area with a color gradient. This represents the color transfer function (or color map), which maps field values to colors.
Before we begin to adjust transfer functions, it is a good idea to set the color map range. This is the range of field values that will be represented in the transfer functions. Values below and above this range will simply be assigned the first and last value in the transfer functions, respectively. By default, the color map range is the full range of values present in the field. If we want to want to focus on a smaller range of values, we can specify the range by clicking the Rescale to custom range button (green double arrow to the left of the transfer function view, second from the top) and specify a new range. For this example, let us specify an upper limit of 2 MK.
Next, we’ll tweak the opacities. Let’s say we want to visualise the structure of the transition region. Grid cells with temperatures of 2 MK and above are not really interesting for this purpose, and only serve to block the view of the transition region. By dragging the right end node of the opacity transfer function (in the upper right) from 1 to 0, we can hide those pesky hot regions. Unfortunately, this collapses the whole linear function and hides everything else as well. We can fix this by adding a new node by clicking anywhere in the opacity transfer funciton view, and dragging it to give more opacity to the temperatures we are interested in. Feel free to play around by adjusting ranges, adding and moving nodes, or even freehand drawing a transfer function. If you mess things up too much, scroll to the bottom of the editor and click the “recycle” button to restore the default transfer functions.